The bootstrap method - Solutions
Data
For this lab, we will use the dataset sphagnum_cover.csv, from the paper:
Maanavilja, L., Kangas, L., Mehtätalo, L. and Tuittila, E.‐S. (2015), Rewetting of drained boreal spruce swamp forests results in rapid recovery of Sphagnum production. J Appl Ecol, 52: 1355-1363. doi:10.1111/1365-2664.12474)
This dataset contains measurements of the percentage cover of sphagnum moss (sphcover) for 36 boreal swamps divided into three types (habitat): Dr = drained, Re = rewetted et Un = undrained.
cover <- read.csv("sphagnum_cover.csv")
str(cover)## 'data.frame': 36 obs. of 3 variables:
## $ site : chr "AmLuxx" "EvLuPa" "EvLuVK" "HeLuxx" ...
## $ habitat : chr "Un" "Un" "Un" "Un" ...
## $ sphcover: num 35.3 56.2 46.6 56 54.3 ...1. Estimation of mean cover for drained swamps
- From the dataset, extract the sphcover values for drained swamps. Calculate the mean percentage cover and its standard error from the usual formula (based on standard deviation and sample size). Finally, calculate the 95% confidence interval based on the \(t\) distribution:
\[(\bar{x} + t_{(n-1)0.025} s_{\bar{x}}, \bar{x} + t_{(n-1)0.975} s_{\bar{x}})\]
Reminder: The function qt(p, df) gives the quantile corresponding to a given cumulative probability p, for a \(t\) distribution with df degrees of freedom.
Solution
cov_dr <- cover$sphcover[cover$habitat == "Dr"]
n <- length(cov_dr)
# Mean
moy <- mean(cov_dr)
moy## [1] 7.16229# Standard error
err_type <- sd(cov_dr) / sqrt(n)
err_type## [1] 2.517416# Confidence interval
ic <- c(moy + qt(0.025, n - 1) * err_type,
moy + qt(0.975, n - 1) * err_type)
ic## [1] 1.357118 12.967461- Simulate 10,000 bootstrap samples for the mean calculated in a). What is its standard error according to the bootstrap? Does this statistic appear biased?
Solution
library(boot)
set.seed(5612)
calc_moy <- function(x, i) mean(x[i])
boot_moy <- boot(cov_dr, calc_moy, R = 10000)
# Erreur-type
sd(boot_moy$t)## [1] 2.386784# Biais
mean(boot_moy$t) - boot_moy$t0## [1] 0.0328031The bias is quite small (compared to the standard error) and could be due to a numerical error rather than a real bias in the statistic.
- How does the bootstrap distribution differ from a normal distribution? To answer this question, it may be useful to draw a quantile-quantile plot (in the code below,
resis the result of the bootstrap):
qqnorm(res$t)
qqline(res$t)Solution
qqnorm(boot_moy$t)
qqline(boot_moy$t)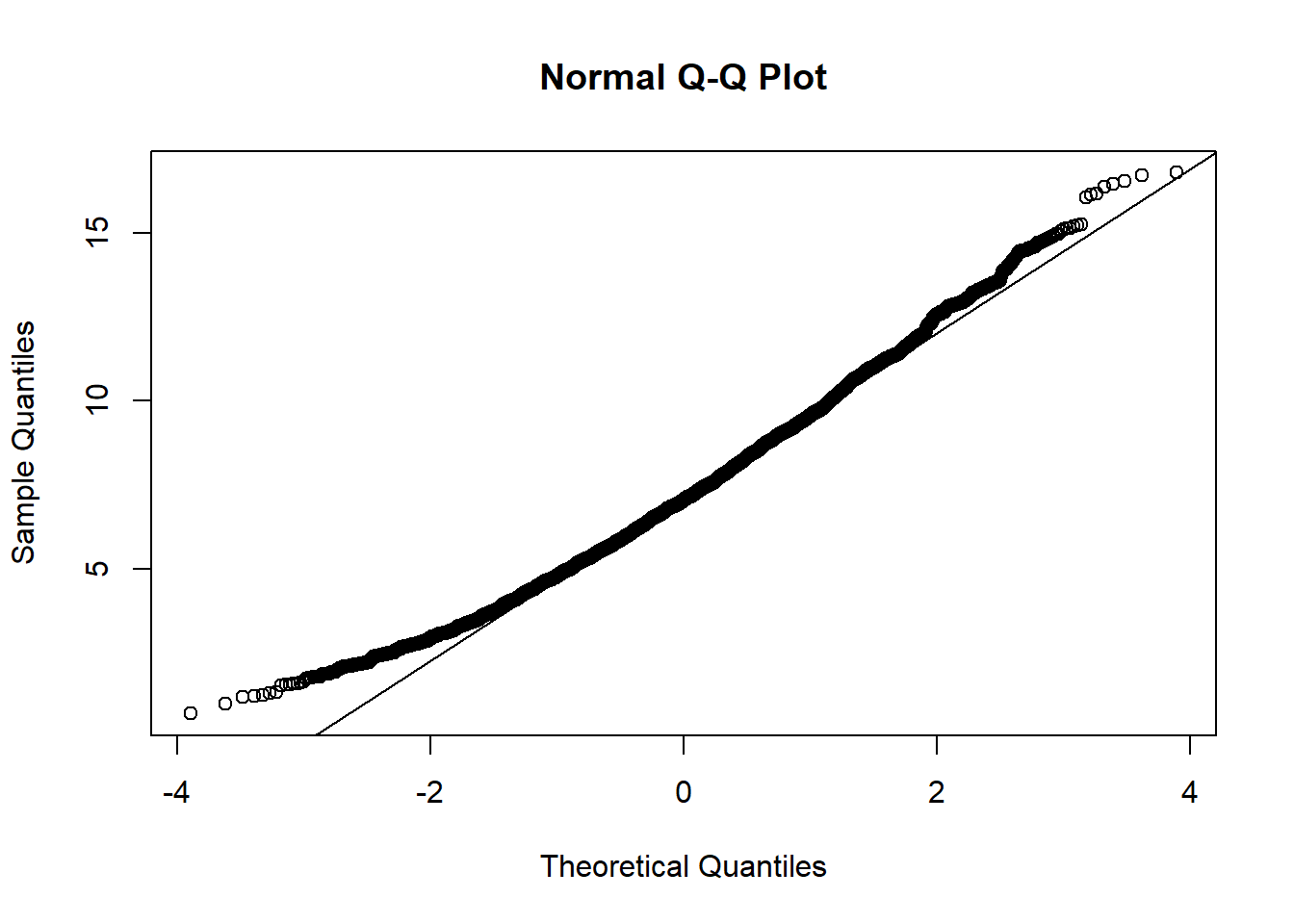
The bootstrap distribution is skewed: the smallest values are above the line, therefore closer to the mean compared to a normal distribution, while the largest values are also above the line, therefore further from the mean.
- Calculate the 95% confidence interval of the mean using the BCa method. How does it differ from the one calculated in a) using the classical formula? Can you explain this difference based on the result in c)?
Solution
boot.ci(boot_moy)## Warning in boot.ci(boot_moy): bootstrap variances needed for studentized
## intervals## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 10000 bootstrap replicates
##
## CALL :
## boot.ci(boot.out = boot_moy)
##
## Intervals :
## Level Normal Basic
## 95% ( 2.451, 11.807 ) ( 1.985, 11.324 )
##
## Level Percentile BCa
## 95% ( 3.001, 12.340 ) ( 3.430, 13.281 )
## Calculations and Intervals on Original ScaleThe BCa interval (3.43, 13.28) differs from that in a) (1.36, 12.97) especially at the lower bound, and both bounds are corrected upwards. In c), we have seen that the smallest values of the bootstrap distribution are closer to the mean with respect to the normal distribution, whereas the largest values are farther away from it.
2. Estimation of differences between habitats
- Here is the distribution of sphcover values in each habitat type.
library(ggplot2)
ggplot(cover, aes(x = habitat, y = sphcover)) +
geom_boxplot()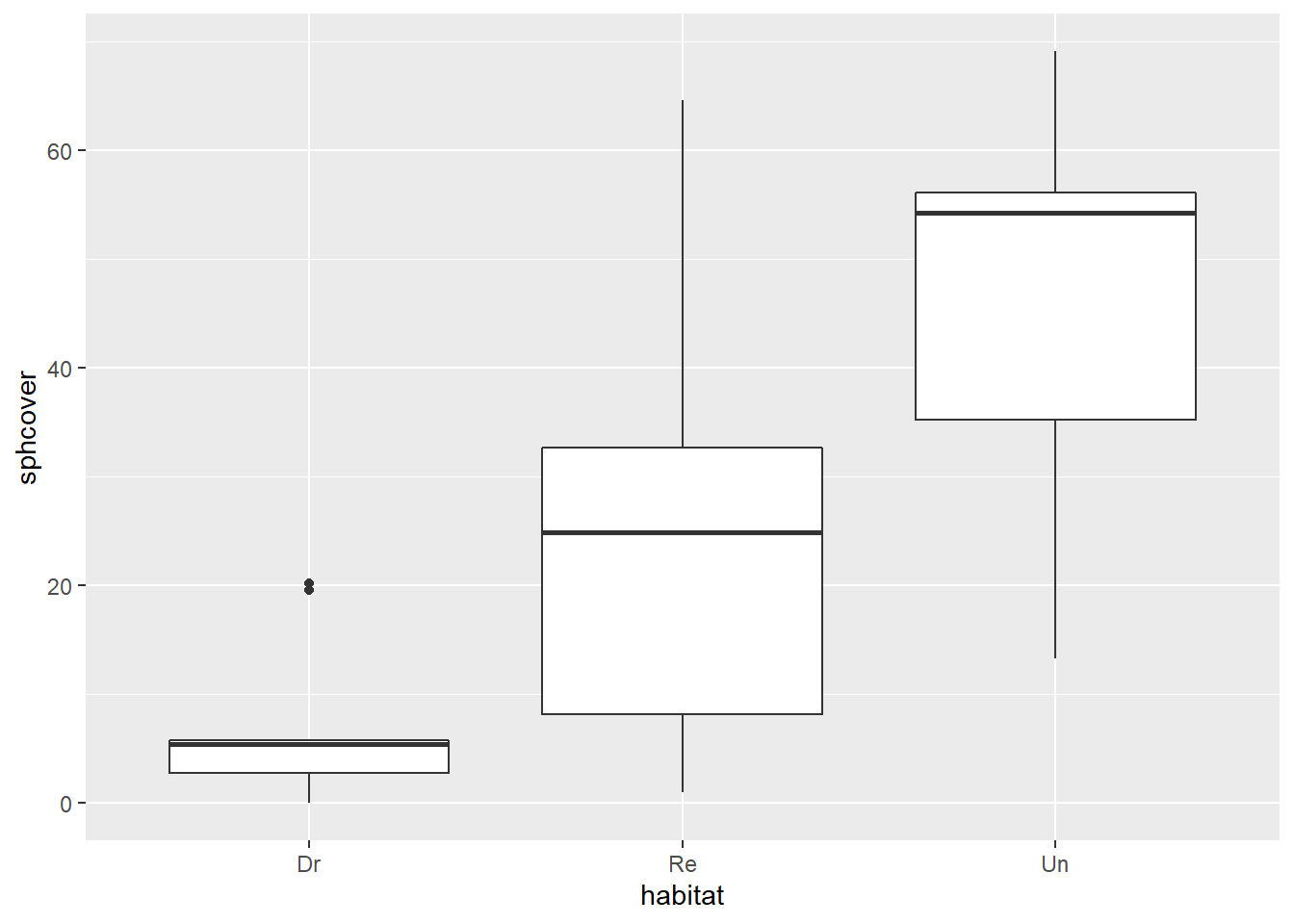
What are the assumptions of a classical ANOVA model that would describe sphagnum moss cover by habitat type? Do these assumptions appear to be met here?
Solution
The ANOVA assumes that the residuals follow a normal distribution of the same variance for each group. Here, the distributions are skewed and the variance seems smaller for the Dr group seems than the others.
- Fit the
sphcover ~ habitatlinear model to thecoverdataset. See the summary of the model results with thesummaryfunction and the confidence intervals of the coefficients with theconfintfunction. What is the interpretation of each coefficient? Are the confidence intervals plausible?
Solution
mod <- lm(sphcover ~ habitat, data = cover)
summary(mod)##
## Call:
## lm(formula = sphcover ~ habitat, data = cover)
##
## Residuals:
## Min 1Q Median 3Q Max
## -33.144 -8.161 -0.596 9.659 41.371
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 7.162 5.129 1.396 0.1719
## habitatRe 16.141 6.282 2.569 0.0149 *
## habitatUn 39.266 7.254 5.413 5.45e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 15.39 on 33 degrees of freedom
## Multiple R-squared: 0.4742, Adjusted R-squared: 0.4424
## F-statistic: 14.88 on 2 and 33 DF, p-value: 2.473e-05The intercept is the mean cover for the reference group (drained swamps, Dr). The coefficient habitatRe (respectively, habitatUn) is the difference between the mean cover of rewetted and drained swamps (respectively, the difference between undrained and drained swamps).
confint(mod)## 2.5 % 97.5 %
## (Intercept) -3.273495 17.59807
## habitatRe 3.360128 28.92248
## habitatUn 24.507522 54.02438The confidence interval for the mean of the Dr group reaches negative values, which is not possible for a percentage cover.
- Create a function with arguments
xandi, which fits the linear model in b) by replacing the original data set (data = cover) withdata = x[i, ], and then returns the coefficients of the model with the functioncoef. Then, applybootto thecoverdataset with the function created and perform 10,000 replicates.
Notes
When the first argument of `boot’ is a dataset, it is the rows of that dataset that are resampled.
Since the statistic calculated by the function has several values (each of the coefficients), the
telement of thebootresult is a matrix rather than a vector. The columns of this matrix correspond to each of the coefficients in order. You can calculate a statistic for each column with the functionapply, e.g.apply(res$t, 2, mean). Here,2indicates to calculate themeanfunction per column (1would mean per row).
Solution
lm_hab <- function(x, i) {
mod <- lm(sphcover ~ habitat, x[i,])
coef(mod)
}
boot_hab <- boot(cover, lm_hab, R = 10000)
# Mean
apply(boot_hab$t, 2, mean)## [1] 7.193903 16.132949 39.288069# Standard error
apply(boot_hab$t, 2, sd)## [1] 2.529526 4.599182 6.523183The mean of the bootstrap is very close to the coefficients estimated in b), but the standard errors have decreased.
Note that all coefficients depend on the mean of the reference group Dr. The mean and standard deviation of this group are strongly affected by two extreme values around 20, as shown in graph a). These extreme values will be absent in several bootstrap samples, which has the effect of reducing the estimated standard deviation for the Dr group and thus reducing the standard error of the model’s coefficients.
- The application of the bootstrap in c) resamples across all rows, so that the number of observations in each habitat type varies from sample to sample. If it is preferable to consider these numbers as fixed quantities, habitat types can be defined as strata by adding the argument
strata = as.factor(cover$habitat)to thebootfunction. (The conversion of the variablehabitatto a factor is necessary here.)
Repeat the analysis in c) with stratified resampling and compare the standard errors obtained for each coefficient.
Solution
boot_strat <- boot(cover, lm_hab, R = 10000, strata = as.factor(cover$habitat))
# Mean
apply(boot_strat$t, 2, mean)## [1] 7.165948 16.180594 39.323302# Standard error
apply(boot_strat$t, 2, sd)## [1] 2.372362 4.490822 6.173863All standard errors have decreased compared to c).
- Calculate the confidence interval for the
habitatUncoefficient according to the result of the bootstrap in d). Note that you need to add the argumentindex = 3to the functionboot.cito tell R to calculate the interval for the 3rd coefficient.
Solution
boot.ci(boot_strat, index = 3)## Warning in boot.ci(boot_strat, index = 3): bootstrap variances needed for
## studentized intervals## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 10000 bootstrap replicates
##
## CALL :
## boot.ci(boot.out = boot_strat, index = 3)
##
## Intervals :
## Level Normal Basic
## 95% (27.11, 51.31 ) (27.86, 51.77 )
##
## Level Percentile BCa
## 95% (26.76, 50.68 ) (25.24, 49.55 )
## Calculations and Intervals on Original Scale- Finally, we will resample the model residuals.
Fit a linear model as in b), then add to the dataset
covera column for the model’sfittedvalues.Write a function that creates a new dataset by adding a resampled vector
x[i]to the expected values to produce a new response variable, then fits a model with this new response variable to the habitat.Simulate 10,000 samples with the
bootfunction, using as arguments (1) the residuals vector (residuals) of the model and (2) the function created above. Do not specify strata. Recalculate the mean, standard error and 95% confidence interval of the coefficients.
Is resampling the residuals a good choice for these data?
Solution
mod <- lm(sphcover ~ habitat, data = cover)
cover$fitted <- fitted(mod)
lm_resid <- function(x, i) {
cover$cover_new <- cover$fitted + x[i]
mod_new <- lm(cover_new ~ habitat, data = cover)
coef(mod_new)
}
boot_resid <- boot(residuals(mod), lm_resid, R = 10000)
# Mean
apply(boot_resid$t, 2, mean)## [1] 7.155993 16.157210 39.340901# Standard error
apply(boot_resid$t, 2, sd)## [1] 4.941989 6.059448 6.965119boot.ci(boot_resid, index = 3)## Warning in boot.ci(boot_resid, index = 3): bootstrap variances needed for
## studentized intervals## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 10000 bootstrap replicates
##
## CALL :
## boot.ci(boot.out = boot_resid, index = 3)
##
## Intervals :
## Level Normal Basic
## 95% (25.54, 52.84 ) (25.71, 52.94 )
##
## Level Percentile BCa
## 95% (25.59, 52.82 ) (25.23, 52.53 )
## Calculations and Intervals on Original ScaleResampling of the residuals assumes that the residuals in each group are from the same distribution. In this case, the variances are not homogeneous between groups, so it is preferable to re-sample the observations.